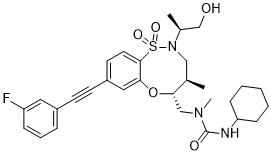
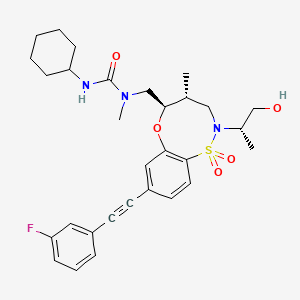
BRD2879
BRD-K56962879-001-01-5
CAS 1304750-47-7
Chemical Formula: C30H38FN3O5S
Molecular Weight: 571.7084
CAS 1304750-47-7
Chemical Formula: C30H38FN3O5S
Molecular Weight: 571.7084
3-cyclohexyl-1-(((4R,5R)-8-((3-fluorophenyl)ethynyl)-2-((S)-1-hydroxypropan-2-yl)-4-methyl-1,1-dioxido-2,3,4,5-tetrahydrobenzo[b][1,4,5]oxathiazocin-5-yl)methyl)-1-methylurea
3-cyclohexyl-1-[[(4R,5R)-8-[2-(3-fluorophenyl)ethynyl]-2-[(2S)-1-hydroxypropan-2-yl]-4-methyl-1,1-dioxo-4,5-dihydro-3H-6,1$l^{6},2-benzoxathiazocin-5-yl]methyl]-1-methylurea
BRD2879 is a potent and cell-active inhibitor of IDH1-R132H with a markedly different structure from previously reported probes with (IC50 = 50 nM for inhibiting IDH1-R132H enzyme). BRD2879 represents a new structural class of mutant IDH1 inhibitors that, with optimization, may prove useful in the study of this enzyme and its role in cancer


PAPER

Evidence suggests that specific mutations of isocitrate dehydrogenases 1 and 2 (IDH1/2) are critical for the initiation and maintenance of certain tumor types and that inhibiting these mutant enzymes with small molecules may be therapeutically beneficial. In order to discover mutant allele-selective IDH1 inhibitors with chemical features distinct from existing probes, we screened a collection of small molecules derived from diversity-oriented synthesis. The assay identified compounds that inhibit the IDH1-R132H mutant allele commonly found in glioma. Here, we report the discovery of a potent (IC50 = 50 nM) series of IDH1-R132H inhibitors having 8-membered ring sulfonamides as exemplified by the compound BRD2879. The inhibitors suppress (R)-2-hydroxyglutarate production in cells without apparent toxicity. Although the solubility and pharmacokinetic properties of the specific inhibitor BRD2879 prevent its use in vivo, the scaffold presents a validated starting point for the synthesis of future IDH1-R132H inhibitors having improved pharmacological properties.
Discovery of 8-Membered Ring Sulfonamides as Inhibitors of Oncogenic Mutant Isocitrate Dehydrogenase 1
† Center for the Science of Therapeutics, Broad Institute, Cambridge, Massachusetts 02142, United States
‡ Department of Chemistry and Chemical Biology, Harvard University, Cambridge, Massachusetts 02138, United States
§ Howard Hughes Medical Institute, Cambridge, Massachusetts 02138, United States
∥ Department of Neurological Surgery, Washington Univeristy School of Medicine, St. Louis, Missouri 63110, United States
ACS Med. Chem. Lett., 2016, 7 (10), pp 944–949
DOI: 10.1021/acsmedchemlett.6b00264
Publication Date (Web): August 18, 2016
Copyright © 2016 American Chemical Society


CLICK ON IMAGES
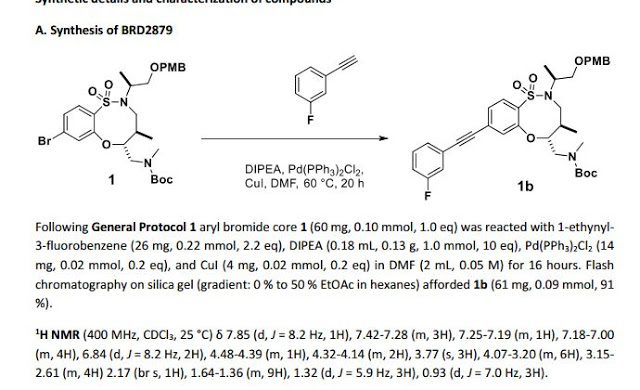
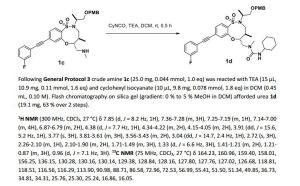
1H NMR (300 MHz, CDCl3, 27 °C) δ 7.88 (d, J = 8.3 Hz, 1H), 7.39-7.27 (m, 3H), 7.22-7.14 (m, 2H), 7.12-7.03 (m, 1H), 4.57 (td, J = 9.5, 2.5 Hz, 1H), 4.35-4.23 (m, 2H), 3.97-3.78 (m, 2H), 3.74-3.60 (m, 2H), 3.51 (ddd, J = 12.3, 9.9, 4.0 Hz, 1H), 3.40 (dd, J = 15.8, 5.1 Hz, 1H), 3.18 (dd, J = 9.9, 2.9 Hz, 1H), 3.12 (dd, J = 14.4, 2.6 Hz, 1H), 2.66 (s, 3H), 2.30-2.16 (m, 1H), 2.06 (d, J = 12.2 Hz, 1H), 1.96 (d, J = 12.1 Hz, 1H), 1.69-1.58 (m,1H), 1.58-1.45 (m, 2H), 1.23 (d, J = 6.8 Hz, 3H), 1.40-0.97 (m, 5H), 0.94 (d, J = 7.0 Hz, 3H).
13C NMR (75 MHz, CDCl3, 27 °C) δ 164.20, 160.92, 157.74, 154.80, 134.60, 130.26, 130.15, 129.56, 128.62, 127.82, 127.77, 127.68, 126.90, 124.27, 118.81, 118.50, 116.63, 116.35, 91.32, 88.40, 85.60, 64.86, 58.03, 51.64, 49.88, 48.51, 36.73, 34.51, 34.35, 34.31, 25.72, 25.28, 25.23, 15.76, 15.05.
HRMS (ESI) calc’d for C30H38FN3O5S [M+H]+ : 572.2589. Found: 572.2588.
1H NMR PREDICT


13C NMR PREDICT

REFERENCES
Discovery of 8-Membered Ring Sulfonamides as Inhibitors of Oncogenic Mutant Isocitrate Dehydrogenase 1
Jason M. Law, Sebastian C. Stark, Ke Liu, Norah E. Liang, Mahmud M. Hussain, Matthias Leiendecker, Daisuke Ito, Oscar Verho, Andrew M. Stern, Stephen E. Johnston, Yan-Ling Zhang, Gavin P. Dunn, Alykhan F. Shamji, and Stuart L. Schreiber
Publication Date (Web): August 18, 2016 (Letter)
DOI: 10.1021/acsmedchemlett.6b00264
Jason M. Law, Sebastian C. Stark, Ke Liu, Norah E. Liang, Mahmud M. Hussain, Matthias Leiendecker, Daisuke Ito, Oscar Verho, Andrew M. Stern, Stephen E. Johnston, Yan-Ling Zhang, Gavin P. Dunn, Alykhan F. Shamji, and Stuart L. Schreiber
Publication Date (Web): August 18, 2016 (Letter)
DOI: 10.1021/acsmedchemlett.6b00264
/////////////2-hydroxyglutarate, allele-selective probe, AML, BRD 2879, cancer, diversity-oriented synthesis, glioma, high-throughput screening, isocitrate dehydrogenase, small-molecule probe, BRD2879; BRD-2879; BRD 2879
FC1=CC(C#CC2=CC(O[C@@H](CN(C(NC3CCCCC3)=O)C)[C@H](C)CN([C@@H](C)CO)S4(=O)=O)=C4C=C2)=CC=C1