
CAPREOMYCIN General Structure
Capreomycin is a peptide antibiotic, commonly grouped with the aminoglycosides, which is given in combination with other antibiotics for MDR-tuberculosis. Adverse effects include nephrotoxicity and 8th cranial auditory vestibular nerve nerve toxicity.
The drug should not be given with streptomycin or other drugs that may damage the auditory vestibular nerve. Patients on this drug will often require audiology tests.
It is a cyclic peptide. Capreomycin is administered intramuscularly and shows bacteriostatic activity.REF 20
Capreomycin is frequently used to treat Mycobacterium tuberculosis infections. Mycobacterium tuberculosis growth has been found to be inhibited at a concentration of 2.5 μg/mL. REF21

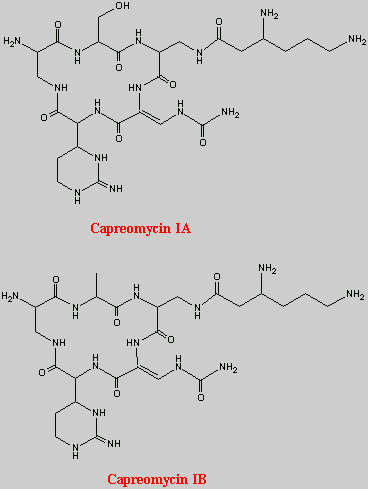
This is the basic structure of capreomycin. The table below identifies the various naturally occuring analogues12, 14.
R1
|
R2
| |
| Capreomycin IA | OH | b-Lys |
| Capreomycin IB |
H
| b-Lys |
| Capreomycin IIA |
OH
| NH2 |
| Capreomycin IIB |
H
| NH2 |

Introduction
Capreomycin is a metabolite of Streptomyces capreolus, it is an antimycobacterial agent - and a potent tuberlostatic antibiotic. Capreomycin is effective against a number of Gram-positive and Gram-negative organisms, but is primarily active against mycobacteria. It has been used in the treatment of certain resistant strains of Mycobacterium tuberculosis. The drug was first described in 1960 be Herr, and was subsequently found to contain two components (I and II) and later to be comprised of four (IA, IB, IIA, IIB) as shown on thestructure page.
Tuberculosis
Tuberculosis is a disease of the respiratory system, and is airbourne. The bacilli implant themselves in areas such as the lungs, renal cortex and reticuloendothelial system where there is a high partial pressure of oxygen. This is the Primary infection and does not normally affect the person whilst their immune system is intact as the bacteria lie dormant. When the immune system is depressed, the secondary reactivation occurs, and effects of the disease are seen.
This infectious disease has been known since about 1000B.C., and it stills remains the 'leading cause of death from a single infectious disease agent'7. It is estimated that around eight million people contract TB every year, of which 95% are in developing countries. Deaths from the disease is estimated at 3 million people per year by the World Health Organisation. The occurance of the disease is related directly to the economic state of the country. This is because the spread of the disease is greatly assisted by poor public and personal hygiene and by overcrowding. New drugs were develoed about forty years ago allowing tuberculosis to be regarded as a curable disease. This is no longer the case, as many multidrug-resistant strains of the disease have emerged. This is where capreomycin has it uses.
There are three groups of drugs used to treat TB, which vary in their effectiveness and potential side effects.
First line drugs include: isoniazid, rifampicin and pyrazinamide. These are most effective and have the fewest potential side effects.
Second line drugs include: ethambutol, streptomycin and p-amino salicyclic acid. These are less effective and have more toxic effects.
Third line drugs include: Capreomycin, cycloserine, viomycin, kanamycin and amikacin. These are least effective and have the most toxic effects.
The third line drugs have to be used for infections with tubercle bacilli, likely to be resistant to first and second line drugs or when first and second line drugs have been abandoned because of unwanted reactions. To decrease the possibility of resistant organisms from emerging, 'Compound Drug Therapy' is used where a concoction of several drugs is administered.
General Physical Data
Molecular Weight | 653.70 |
| Molecular Formula | C25H43N13O8 |
| CAS Registry number | 61394-77-2 |
| Beilstein Registry number | 876587 |
| Chemical Name |
L-3,6-diamino-hexanoyl->-cyclo-[L-2,3-diamino-propionyl->-L-seryl->-L-alanyl->-2-amino-3-ureido
-acryloyl->-(S)-amino-((R)-2-amino-1(3),4,5,6-tetrahydro-pyrimidin-4-yl)-acetyl-(1->N%3&)]
|
| Auto name |
3,6-diamino-hexanoic acid [12-hydroxymethyl-3-(2-imino-hexahydro-pyrimidin-4-yl)-9-methyl-
2,5,8,11,14-pentaoxo-6-ureidomethylene-1,4,7,10,13-pentaaza-cyclohexadec-15-yl]-amide
|
| Cpm IA10 | Cpm IB10 | Cpm IIA14 | Cpm IIB14 | ||
| m.p. / oC | 240-5 | 250-3 | 250 | 252 | |
| [a]D / o | -22.0 | -42.5 | +9.3 | -24.9 | |
| UV / nm | 0.1 M HCl | 269 (e 23, 400) | 268 (22, 000) | ||
| H2O | 268 (23, 200) | 268 (21, 900) | |||
| 0.1 M NaOH | 288 (15, 800) | 290 (13, 100) | |||
According to the literature9the following applies to naturally occuring capreomycin:
Ratio of IA to IB = 1.16
Capreomycin II = 1.5%
Ratio of IA to IB = 1.16
Capreomycin II = 1.5%
13C NMR Data of Cpm IA

| Carbon Number | d / ppm |
| 1 | 51.92 |
| 2 | 40.28 |
| 4 | 172.76 |
| 10 | 176.29 |
| 11 | 54.15 |
| 5, 14 | 55.66 |
| 56.23 | |
| 7 | 168.0 |
| 8 | 105.90 |
| 13 | 172.00 |
| 16 | 176.6 |
| 17 | 135.79 |
| 19 | 155.32 |
| 20 | 18.86 |
| 21 | 68.33 |
| 22 | 49.20 |
| 23 | 23.53 |
| 24 | 49.83 |
| 26 | 157.0 (b) |
| 1' | 172.0 |
| 2' | 36.93 |
| 3' | 49.26 |
| 4' | 23.59 |
| 5' | 29.77 |
| 6' | 39.77 |
The included NMR data is taken from tables in the literature8, 14
The 13C NMR data is that of Capreomycin IA only, and the carbons are numbered accordingly in red on the structure shown above.
Below are 1H NMR tables for the four different naturally occurring forms of capreomycin, the NH protons and CH protons are given in different tables. The NH protons are again numbered on the Cpm IA structure above, but this time in blue. The CH protons are numbered according to their postion in the amino acid residue. These are also numbered in pink on the above diagram.

Chemical Shifts of CH protons in Capreomycin Analogues

Properties:Crystals. Mp: 246–248°C.
Synonyms:capreomycin IA;Cyclo[A2pr*-Ser-N3-[(3S)-3,6-diamino-1-oxohexyl]A2pr-2-[(Z)-aminocarbonylaminomethylene]Gly-2-[(4R)-2-iminohexahydropyrimidine-4-yl]Gly-]
Synthesis









Capreomycin



| Position of Amino Acid Residue | Cpm IA | Cpm IB | Cpm IIA | Cpm IIB | |
| 1 | a-CH2 | 2.63 (dd) | 2.5 (dd) | ||
| 2.85 (dd) | 2.81 (dd) | ||||
| b-CH2 | 3.8 (m) | 3.7 (m) | |||
| g-CH2 | 1.8 (m) | 1.8 (m) | |||
| d-CH2 | 1.8 (m) | 1.8 (m) | |||
| e-CH2 | 3.10 (m) | 3.08 (m) | |||
| 2 | a-CH | 4.3-3.5 (m) | 4.2-4.5 (m) | 4.3-4.6 (m) | 4.3-4.6 (m) |
| b-CH2 | 3.3 (m) | 3.3 (m) | 3.3 (m) | 3.3 (m) | |
| 3.8 (m) | 3.8 (m) | 4.1 (m) | 4.1 (m) | ||
| 3 | a-CH | 4.86 (t) | 4.67 (q) | 4.84 (t) | 4.68 (q) |
| b-CH2 | 3.84 (d) | 3.95 (d) | |||
| b-CH3 | 1.43 (d) | 1.45 (d) | |||
| 4 | a-CH | 4.3-4.5(m) | 4.2-4.5 (m) | 4.3-4.5 (m) | 4.3-4.5 (m) |
| b-CH2 | 3.7-4.2 (m) | 3.7-4.2 (m) | 3.7-4.2 (m) | 3.79 (dd) | |
| 3.8-4.2 (m) | |||||
| 5 | b-CH | 8.04 (s) | 8.03 (s) | 8.05 (s) | 8.04 (s) |
| 6 | a-CH | 5.01 (d) | 4.96 (d) | 5.01 (d) | 4.95 (d) |
| b-CH | 4.5 (m) | 4.5 (m) | 4.5 (m) | 4.5 (m) | |
| g-CH2 | 1.6-2.3 (m) | 1.6-2.3 (m) | 1.6-2.3 (m) | 1.6-2.3 (m) | |
| d-CH2 | 3.3 (m) | 3.3 (m) | 3.3 (m) | 3.3 (m) |
Chemical Shifts of NH Protons of Capreomycin Analogues
| Cpm IA | Cpm IB | Cpm IIA | Cpm IIB | |
| 1 | 9.33 (d) | 9.72(d) | 9.60 (d) | 9.50 (d) |
| 2 | 9.24 (d) | 9.24 (d) | 9.33 (d) | 9.30 (d) |
| 3 | 8.82 (s) | 8.76 (s) | 9.10 (s) | 9.10 (s) |
| 4 | 8.64 (d) | 8.68(d) | 8.73 (d) | 8.73 (d) |
| 6 | 8.22 (t) | 8.15 (t) | ||
| 7 | 8.10 (t) | 8.15 (t) | 8.19 (t) | 8.08 (t) |
| 8 | 7.61 (d) | 7.62 (d) | 7.50 (d) | 7.49 (d) |
| 9 | 7.46 (s) | 7.42 (s) | 7.44 (s) | 7.44 (s) |
| 10 | 7.46 (s) | 7.42 s) | 7.31 (s) | 7.18 (s) |
| 11 | 6.48 (s) | 6.49 (s) | 6.43 (s) | 6.34 (s) |
| 12 | 6.29 (s) | 6.34 (s) | 6.29 (s) | 6.27 (s) |
Using the program gNMR I attempted to plot the above data. However, this was not successful as this program can only cope with molecules with up to 23 protons. As this molecule has Capreomycin IA has 43 hydogens, the generated 1H NMR was lacking many essential peaks, and hence was not included.
IR Spectrum of Capreomycin IA
The same process could have done for any of the other three Capreomycin anlogues. The very broad band around 2000 cm-1 upwards is due to the presence of so many nitrogen and carbonyl groups and hence hydrogen bonding.
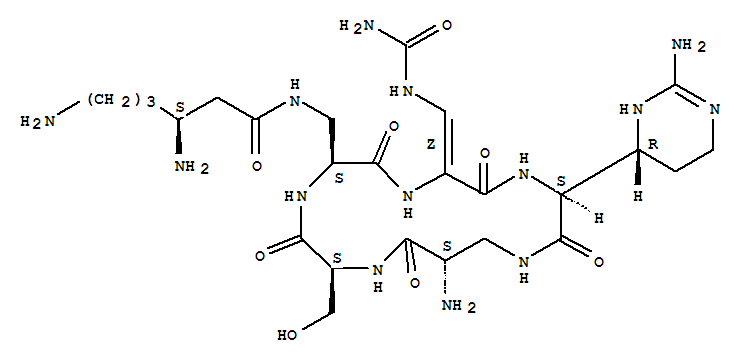
Cyclo[3-[[(3S)-3,6-diamino-1-oxohexyl]amino]-L-alanyl-(2Z)-3-[(aminocarbonyl)amino]-2,3-didehydroalanyl-(2S)-2-[(4R)-2-amino-3,4,5,6-tetrahydro-4-pyrimidinyl]glycyl-(2S)-2-amino-b-alanyl-L-seryl]
capreomycinIA;Cyclo[3-[[(3S)-3,6-diamino-1-oxohexyl]amino]-L-alanyl-(2Z)-3-[(aminocarbonyl)amino]-2,3-didehydroalanyl-(2S)-2-[(4R)-2-amino-1,4,5,6-tetrahydro-4-pyrimidinyl]glycyl-(2S)-2-amino-b-alanyl-L-seryl] (9CI);1,4,7,10,13-Pentaazacyclohexadecane, cyclic peptide deriv.
37280-35-6
| Formula: | C25H44 N14 O8 |
|---|---|
| Molecular Weight: | 668.83 |
Synonyms:capreomycin IA;Cyclo[A2pr*-Ser-N3-[(3S)-3,6-diamino-1-oxohexyl]A2pr-2-[(Z)-aminocarbonylaminomethylene]Gly-2-[(4R)-2-iminohexahydropyrimidine-4-yl]Gly-]
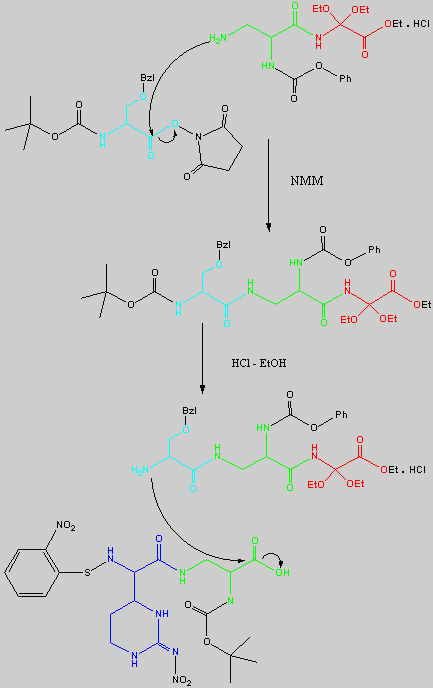
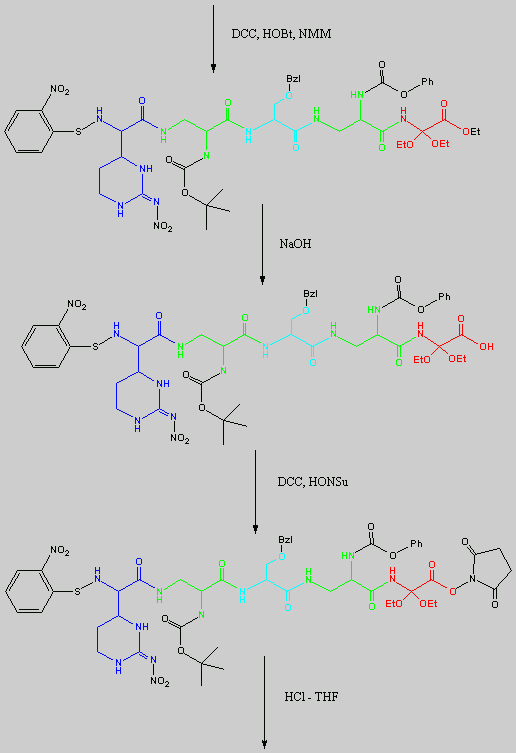
Below is the peptide synthesis of capreomycin IA and IB. This was taken directly from the literature10.


No chemical synthesis of capreomycin could be found in any of the literature references. However, below is a synthesis devised from the peptide synthesis shown above. This is colour coded depending on the various amino residues. Each of the amino groups is added to the molecule in sequence linked by a peptide bond to eventually form the cyclo-structure. This was designed with some help from general references1,2,3.

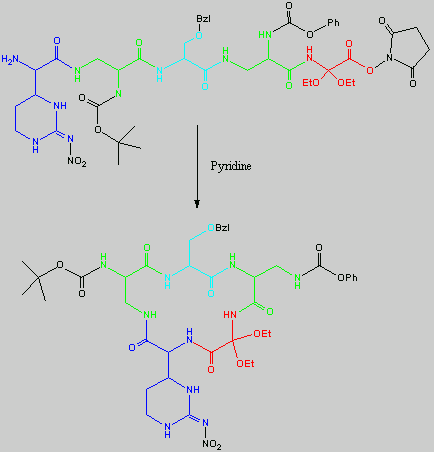
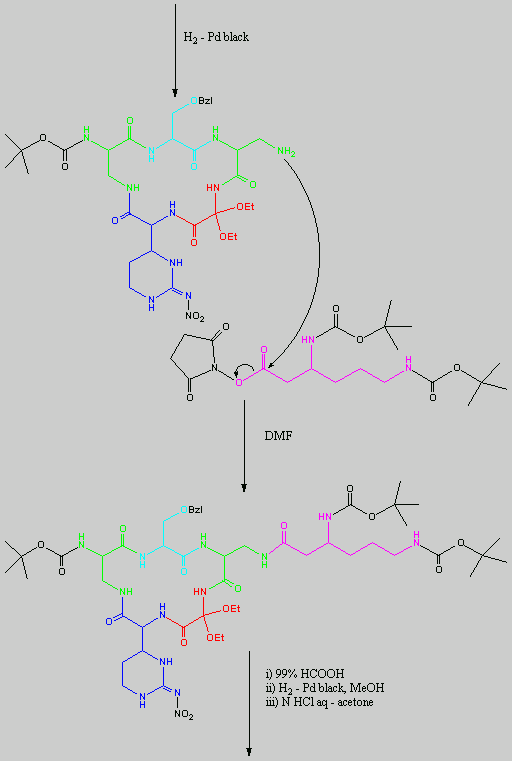
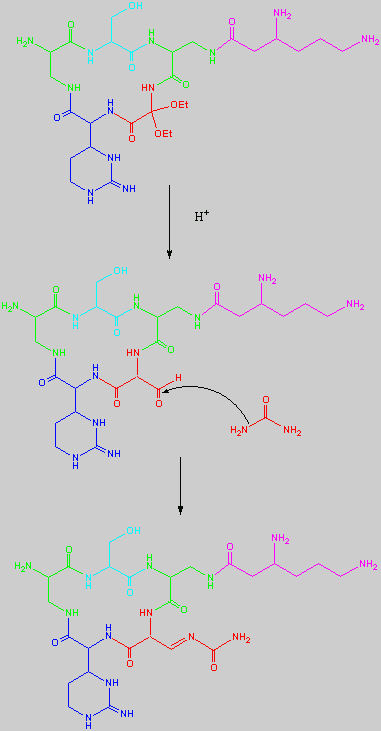
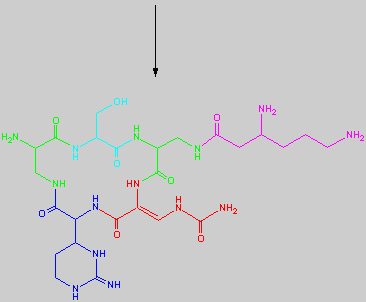
This synthesis would be identical for capreomycin IB other than the Serine-Bzl is replaced by Alanine and the synthesis works in exactly the same way.
The individual components of the capreomycin were colour coded as follows:
| Red | DEA / UDA |
b, b � diethoxyalanine / b - ureidodehydroalanine
|
| Green | A2pr | a, b � diaminopropionic acid |
| Turquoise | Ser | Serine |
| Blue | Cpd | Capreomycidine |
| Pink |
b-Lys
|
b-Lysine
|
The black components of the synthesis were the various protecting groups involved:
| Boc | tert � butoxycarbonyl | 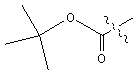 |
| Z | Benzyloxycarbonyl | 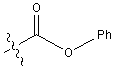 |
| ONSu | N-hydroxysuccinimide | 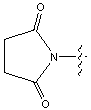 |
| Nps | o-Nitrophenylsulphenyl | 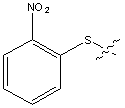 |
NO2
| Nitro |
NO2
|
| Bzl | Benzene |  |
| NMM | N-Methylmorpholine |
| DCC | N,N�-Dicyclohexylcarbodiimine |
| HOBt | l-Hydroxybenztriazole |
| HONSu | N-Hydroxysuccinimide |
| THF | Tetrahydrofuran |
This is the synthesis of capreomycin IA. The IB form is produced in an identical fashion except that Ser � Bzl , is replaced with Ala.
is replaced with Ala. 
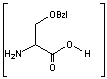 is replaced with Ala.
is replaced with Ala. 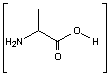
..........................


Capastat Sulfate (capreomycin for injection) is a polypeptide antibiotic isolated from Streptomyces capreolus. It is a complex of 4 microbiologically active components which have been characterized in part; however, complete structural determination of all the components has not been established.
Capreomycin is supplied as the disulfate salt and is soluble in water. In complete solution, it is almost colorless.
Each vial contains the equivalent of 1 g capreomycin activity.
The structural formula is as follows:

Biological Action
Capreomycin is part of a group of drugs called aminoglycosides. These act to inhibit bacterial protein synthesis. The oxygen-dependent active transport by a polyamine carrier system affects the penetration of the aminoglycosides through the cell membrane of the bacterium. Minimal action on anaerobic organisms is observed. The effect of the aminoglycosides is bactericidal and is enhanced by agents that interfere with cell wall synthesis.
Very little is known about the mechanism of action of capreomycin specifically, but it is thought to inhibit protein synthesis by binding to the 70s ribosomal unit. Other sources6support this theory by suggesting that capreomycin "prevents protein biosynthesis by inhibiting group I intron splicing of RNA as well as blocking translation on the bacterial ribosome via inhibition of ribosomal subunits." It has been reported14 that the b-amino group of the A2pr residue promotes biological potency, and that its location within the molecule is of importance.
Side Effects
This powerful antimycobacterial agent can give rise to several side effects, some of which are listed below:
The following Nephrotoxic effects are reversible once treatment is stopped, but capreomycin is not recommended for people with kidney disorders.
- Polyuria (excess urination)
- Haematuria (red blood cells in the urine)
- Proteinuria (protein in the urine)
- Nitrogen metabolism
- Electrolyte disturbances
- Anorexia
- Anaemia
- Thirst
Capreomycin is also Ototoxic giving the following side effects. The nerve damage is permanent.
- Deafness
- Loss of vestibular function
- Damage to the cranial nerve 8
- References:1. An Introduction to Peptide Chemistry - P.D. Bailey
2. Organic Chemistry - Vollhardt and Schore
3. Peptide Synthesis - M. Bodanszky, Y. Klausner and M. Ondetti
4. Pharmacology - H.P. Rand, M.M. Dale and J.M. Ritter
5. http://www.aidsinfonyc.org/network/access/drugs/capr.html
6. http://rwingo1.chm.colostate.edu/group/duane/duane.html
7. http://www.hucmlrc.howard.edu/Pharmacology/handouts/TBRCLSIS.html
8. J. Org.Chem.,1977, 42, 8 - McGahren, Morton, Kunstmann, Ellestad
9. Bull.W.H.O., 1972, 47(3), 343-56 - Lightbrown et al.
10. Tetrahedron, 1978, 34(7), 912-7 - Nomoto, Teshima, Wakamiya, Shiba
11. Tetrahedron Letters, 1976, 43, 3907-10 - Shiba, Nomoto, Teshima, Wakamiya
12. J.Org.Chem., 1992, 57, 5214-5217 - Gould and Minott
13. Tetrahedron Letters, 1969, 30, 2549-41 - Bycroft, Cameron, Hassanali-Walji and Johnson
14. Bull.Chem.Soc.Jpn, 1979, 52(6), 1709-15 - Nomoto and Shiba
15. Experimentia - 1976, 32(9), 1109-11 - Nomoto and Wakamiya
16. Pharmazie - 1970, 25(8), 471-2 - Voigt and Maa Bared
17. Antimicrobial Agents Chemotherapy, 1964, 522-9 - Black, Griffith and Brickler
18. Antimicrobial Agents Chemotherapy, 1962, 201-12 - Herr
19. www2.chemie.uni-erlangen.de/services/telespec - 20 "Capreomycin binds across the ribosomal subunit interface using tlyA-encoded 2'-O-methylations in 16S and 23S rRNAs". Mol. Cell 23 (2): 173–82. July 2006. doi:10.1016/j.molcel.2006.05.044. PMID 16857584
- 21 http://www.toku-e.com/Assets/MIC/Capreomycin%20sulfate.pdf
- CAPREOMYCIN wiki
 | |
|---|---|
 | |
| Systematic (IUPAC) name | |
| (3S)-3,6-diamino-N-[[(2S,5S,8E,11S,15S)-15-amino-11-[(4R)-2-amino-3,4,5,6-tetrahydropyrimidin-4-yl]-8-[(carbamoylamino)methylidene]-2-(hydroxymethyl)-3,6,9,12,16-pentaoxo-1,4,7,10,13-pentazacyclohexadec-5-yl]methyl]hexanamide; (3S)-3,6-diamino-N-[[(2S,5S,8E,11S,15S)-15-amino-11-[(4R)-2-amino-3,4,5,6-tetrahydropyrimidin-4-yl]-8-[(carbamoylamino)methylidene]-2-methyl-3,6,9,12,16-pentaoxo-1,4,7,10,13-pentazacyclohexadec-5-yl]methyl]hexanamide | |
| Clinical data | |
| AHFS/Drugs.com | monograph |
| MedlinePlus | a682860 |
| Identifiers | |
| CAS number | 11003-38-6 |
| Chemical data | |
| Formula | C25H44N14O8 |
| Mol. mass | 668.706 g/mol |